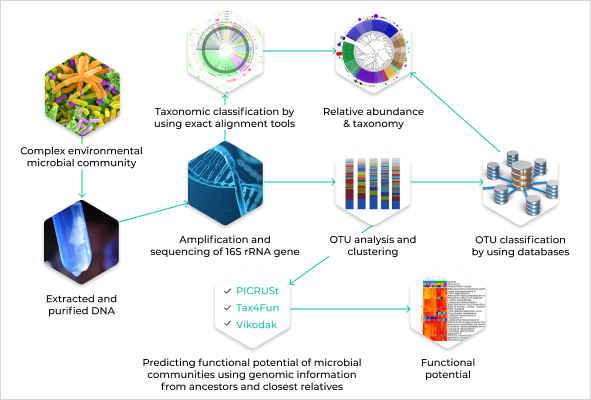
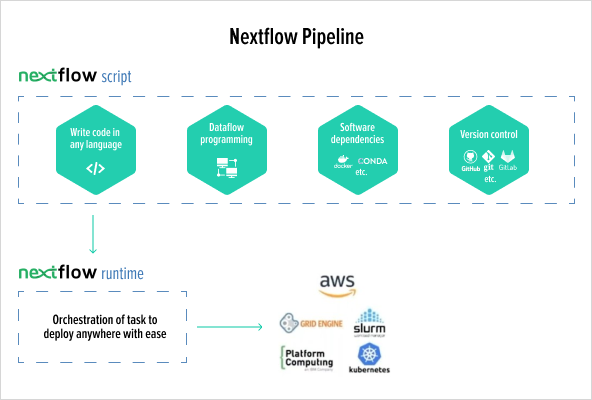
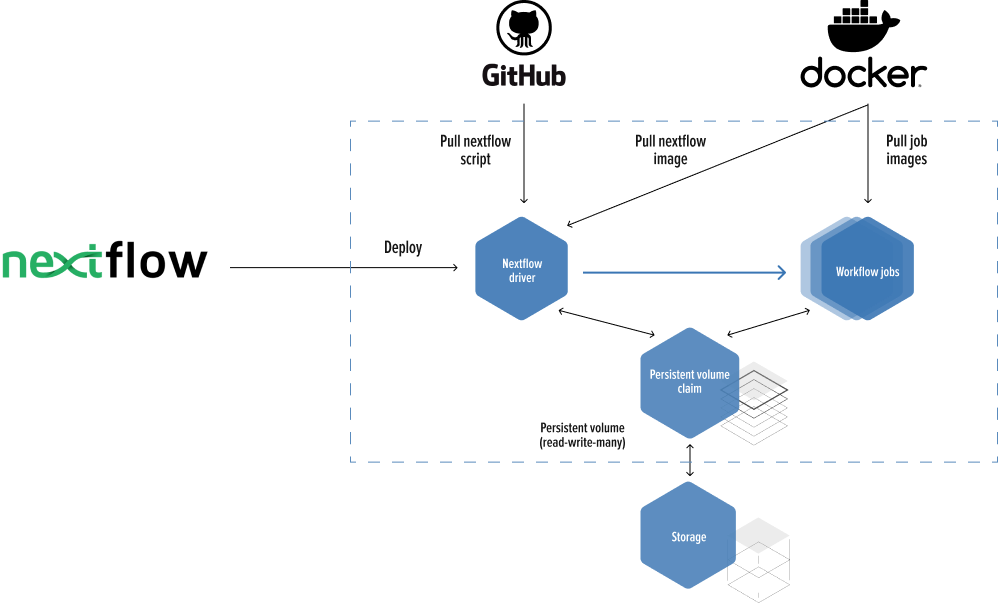
Solution
Migrate an R script from a research article into an NextFlow scalable pipeline running in high performance cloud
Engagement model
Research Partner
Methodology
Agile
Industry
Bioinformatics
Team
BioOps Engineers 1
Bioinformatics Engineer 1
R/Python engineer 1